Amino Acid and Sulfur Metabolism
Cysteine and methionine are two essential amino acids which contain sulfur. We are also looking at interconnections between sulfur metabolism and other plant nutrients. Further, we are investigating means of improving the nutritional quality of crops, with a current focus on rice (Oryza sativa) with respect to a balanced amino acid composition.
In our studies of plant sulfur metabolism, we use two mutually supporting approaches as the basis of our research portfolio. The first is a targeted, pathway-oriented approach aimed at understanding pathway architecture and coordination, and the regulation of the sulfur-containing metabolites as such. The second is a non-biased approach in which functional genomics is used to work out how sulfur metabolism is embedded and controlled within the whole plant system.
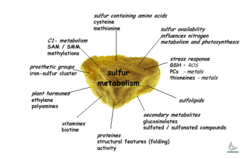
Inorganic sulfate is taken up through plant roots and, via cysteine biosynthesis, incorporated as organic sulfur. Our investigations focus on fundamental questions about cysteine (cys) and methionine (met) biosynthesis and on the possibility of engineering crop plants enriched in these sulfur-containing amino acids. Methionine is essential for non-ruminant mammals (including man) and uptake of cysteine reduces the methionine requirement. We have used transgenic strategies to generate many plant lines affected in cysteine and methionine biosynthesis, and subjected them to detailed molecular and biochemical analyses. Recently, we embarked on a course to study sulfur metabolism in a holistic way, rather than focusing on single pathways as such. By applying functional genomic tools like transcript, metabolite, and protein profiling in our analysis of transgenic potato (Solanum tuberosum) and of the model plant Arabidopsis thaliana, we are heading for a better understanding of the sulfur metabolism network in plants.
To learn about the control mechanisms involved in sulfur-containing amino acid biosynthesis, we are isolating and studying the involved genes and their promoters. The model plant systems of our investigations are potato and Arabidopsis, although a limited amount of work is also dedicated to rice (Oryza sativa), cucumber (Cucumis sativus), and tomato (Lycopersicon esculentum). Various transgenic plants exhibiting reduced or increased expression of relevant genes in the pathway have been produced and analysed. Fundamental knowledge of pathway regulation has been obtained as well as an improvement of the nutritional quality of a crop plant: Nutritional quality is largely determined by methionine, which is often the most limited of the essential amino acids.
The main thrust of our research recently shifted to analysing sulfur metabolism networks. In a systems biology approach, we investigate the response of Arabidopsis to different periods or degrees of sulfur starvation by applying non-biased, multiparallel tools including transcript, protein, and metabolite profiling. Our results are integrated to form working models for further detailed investigations with a focus on regulatory aspects of metabolism. This work entails the detailed analysis of Arabidopsis mutants and pulls many of our earlier results together into biological context (eg. the increased thiol levels seen during SAT over-expression, glutathione involvement in stress response mechanisms towards active oxygen species, etc.). Our long-term goal is to imbed sulfur metabolism in a broader context such as carbohydrate and nitrogen metabolic networks, which will occur through close collaborations with external and in house research groups.





