Applied Metabolome Analysis
Considering the limitations of existing profiling technology from wet-lab experimentation to data visualization and interpretation, the improvement of the core technology, GC-EI-TOF-MS based metabolite profiling, and subsequent data analysis moved to the group focus. Improvement of quantitative accuracy, enhancement of the spectrum of simultaneously monitored metabolites, and developments towards flux phenotyping are top priorities.
Roots are the interface between plant and soil. Root growth is tightly regulated according to the metabolic status of the plant. Thus experiments, which investigate root metabolism, cannot be fully interpreted without insight into the nutrient supply through the shoot and soil substrate. Metabolism itself is highly plastic, meaning that it reacts rapidly to change and can adapt. The multi-parallel gas chromatography- mass spectrometry (GC-MS) technology [1] developed at the MPI-MP since 1998 allows researches to generate profiles, which convey information on the concentration levels of hundreds of known and not yet identified metabolites. We are in the long run interested in developing in silico models for root primary metabolism based on empirical data sets. However, knowledge of the changes in metabolite concentration levels does not suffice. We decided to enhance conventional GC-MS profiling technology towards flux analysis. Thus the focus of the group was moved towards metabolomic and fluxomic technology development utilizing the power of in vivo stable isotope labelling.
Arabidopsis thaliana is the model for developing the stable isotope labelling technology in higher plants (Fig. 2). In parallel conventional high-throughput GC-MS characterization of metabolic changes in response to environmental stimuli, chemical or nutritional supplementation and genetic change is continuously improved and offered in co-operations with guest scientist and in-house partners. The models blue algae and yeast (Saccharomyces cerevisiae) are applied to test and compare experimental concepts, which are difficult to perform in higher plants [2]. Microbial organisms are used to facilitate development of analytical and data mining strategies, especially time course analyses, quantitative standardization and kinetic flux measurements in combination with 13C-stable isotope labelling.
Developing technology to suit the needs

Our group has established and is continuously improving the basic tools for metabolite profiling analyses. Gas chromatography (GC) coupled with electron impact ionization (EI) and time of flight (TOF) mass spectrometry (MS) is the routine technology platform and has been automated through robotic derivatization timed and in line with GC-EI-TOF-MS analysis.
The most essential tool is a collection of frequently measured metabolites and yet non-identified compounds. In co-operation with the in-house bioinformatics group this collection is made available to the public through a data base, the Golm Metabolome Database (GMD ) [3]. The mass spectral tags (MSTs), i.e. mass spectral fragmentation pattern linked to chromatographic retention behaviour, which represent metabolites in GC-MS analyses, are obtained from a collection of approximately 1000 authenticated reference substances. MST libraries are essential, because prior to quantification this information is required for the identification of the metabolites, which are present in GC-MS analyses.
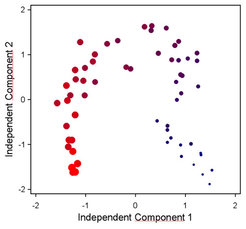
We are also developing data processing, verification, mining and interpretation capabilities, which are adequate for multi-parallel system analyses. For example independent component analysis (ICA) has been established as a routine data visualization tool (Fig. 3) and correlation analysis has been explored using publicly available transcript profiles (cf. CSB.DB).

A software platform, the TagFinder, is used for the quality assessment of GC-MS experiments, chromatographic alignment using retention time standardization, data matrix generation. TagFinder is designed for both, the fingerprinting analyses enabling non-biased marker discovery and the metabolite targeted data retrieval for pathway analysis. The implemented metabolite identification tools use both chemical standards and software support for the matching of retention time behaviour and mass spectral fragmentation.
Exploiting the power of stable isotope labelling
Stable isotope labelling of organisms is one of the most versatile tools of retrieving otherwise non-accessible physiological information from biological systems. Our group uses in vivo 13C-labelling for three essential purposes [2].
(1) Fully 13C-labelled organisms yield metabolic MSTs, which exhibit a mass shift of 1 amu for each - in ambient preparations predominant - 12C-carbon atom that has been replaced, respectively. Thus in vivo feeding of 13C-precursosrs does not only proof the metabolic origin compared to laboratory contaminations, this approach also yields essential information on the carbon composition of yet non-identified metabolites.
(2) The lesson learned from our venture into phytohormone analyses was the absolute requirement of stable isotope labelled internal standardization for trace compound quantification. Thus using the yeast model we now use metabolome extracts from fully 13C-labelled reference material for the relative quantification of each known and non-identified compound utilizing mass isotopomer ratios.
(3) Having labelling capabilities established the next step, which will lead us towards flux analyses in higher plants is obvious. We started to use enclosed and controlled environments in customized growth chambers for the tracing of mass isotopomers under photosynthetically autotrophic conditions. First simultaneous kinetics of 13CO2-incorpoation into root and shoot metabolism of Arabidopsis thaliana have been obtained.
Root Metabolism started up in January 2001 and was modified to Applied Metabolome Analysis / Root Metabolism in 2006.





