Peptide-Protein Interaction Screen
Many regulatory protein-protein interactions are mediated by modified, short, unstructured sequences, which are recognized by specific peptide binding domains (Pawson and Scott 1997). For example, proteins with SH2-domains (Src homology domain 2) bind to phosphorylated tyrosine in a specific sequence context, 14-3-3 proteins recognize a characteristic consensus sequence containing a phosphoserine, and proteins with SH3-domains (Src homology domain 3) bind proline rich peptide motifs. This interplay of protein modification and modification-dependent protein interaction is a key process in regulation of cellular activities (Mayer 2001; Cesareni et al. 2002; Yaffe 2002).
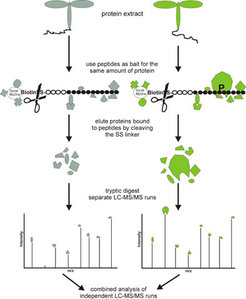
The peptide-protein interaction screen is based on two closely related synthetic bait peptides which are synthesised in an 'active' (e.g. phosphorylated) form and in a 'control' (e.g. non-phosphorylated) form. In separate affinity pull-down experiments, 'active' and 'control' form of the bait peptide are incubated with plant protein extracts. Those proteins that bind to the bait peptides are subsequently eluted. After a tryptic digest, the eluted proteins are identified by mass spectrometry. Tryptic peptides of proteins that specifically bind to the 'active' form of the bait peptide are only identified in the eluate of the pull-down with the 'active' bait peptide, while those proteins that unspecifically bind to both forms of the bait peptids are identified in both eluates (Schulze et al. 2003; Schulze et al. 2005).
The peptide-protein interaction screen provides a powerful tool to identify interaction partners of characteristic phosphoserine or phosphothreonine motifs of receptor-like kinases. We will use this approach to study phosphorylation motifs of nutrient-regulated receptor kinases and also provide first general insights into modification-dependent interaction partners in plant signaling pathways.
Recommended Reading
Pawson T and Scott JD (1997). "Signaling through scaffold, anchoring, and adaptor proteins." Science 278: 2075-2080.
Cesareni G, Panni S, et al. (2002). "Can we infer peptiderecognition specificity mediated by SH3 domains?" FEBS Letters 513: 38-44.
Mayer BJ (2001). "SH3 domains: compexity in moderation." Journal of Cell Science 114(7): 1253-1263.
YaffeMB(2002). "How do 14-3-3 proteins work? - Gatekeeper phosphorylation and the molecular anvil hypothesis." FEBS Letters 513: 53-57.
Schulze W, Mann M (2003) A novel proteomic screen for peptide-protein interactions. Journal of Biological Chemistry: 279: 10756-10764;
Schulze W, Deng L, Mann M (2005) The phosphotyrosine Interactome of the ErbB-receptorfamily. Molecular Systems Biology: doi:10.1038/msb4100012
