Quantitative Genetics
We are currently performing GWAS in Arabidopsis, maize, rice and tomato in order to identify metabolic quantitative trait loci (mQTL). To do so, we are first preforming metabolic-phenotypic characterization of the metabolites (primary and secondary metabolites) and lipids in the different accessions of Arabidopsis, maize, rice and tomato, in different plant tissues, using gas- or liquid-chromatography mass-spectrometry (GC-MS and LC-MS) analysis. The outcome of the GWAS is dense high resolution genetic maps of mQTL. An advantage of the genetic mapping approach is that it is an unbiased approach that allows the simultaneous identification of hundreds of genomic loci that control metabolites and lipid biosynthesis in plants.
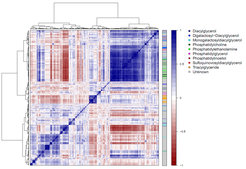
Hierarchical clustering analysis of the lipid-lipid correlation matrix across accessions. Pairwise Pearson correlations (r) among all 722 measured features. Features are ordered using Ward clustering on pairwise dissimilarity, calculated as 1 – r. The cluster on the right is colored according to the chemical class of the lipids.
In Arabidopsis we could identify 309 mQTL for 9 lipid classes, and identified 66 genes in close vicinity to the mapped metabolites, among them more than 30 genes annotated or reported as lipid-metabolism genes. We could validate the function of 25 of those genes in lipid metabolism through the analysis of knockout T-DNA insertion lines of those genes.
In Arabidopsis, we utilize a complementary strategy to support the identification of casual genes by genetic mapping. We integrated mQTL derived from GWAS study of primary metabolites with metabolite‒transcript correlation networks (Wu et al 2016). Low mapping resolution normally limits the discovery to one causal gene per locus; this approach enables us to select candidate genes linked to specific metabolites. We demonstrated that by using an integrative strategy, we can validate previously characterized gene–metabolite associations, and most importantly, identify novel associations between metabolites and genes. This combined quantitative genetics and metabolite‒transcript networks approach can be applied to other organisms and fields of research. We extend this approach using untargeted metabolomics-based GWAS for 2000 semi polar metabolites (mostly secondary), this time with the added value of conducting the GWAS under two distinct environmental conditions, namely heat/darkness stress, and control. The novelty of these two efforts is the usage of environmental stress-driven perturbations of metabolic homeostasis to find novel metabolite-gene associations, some unique to certain environments. (Studied by Si Wu).
Rice is another plant species for which we are currently constructing mQTL maps. We are conducting a GWAS using grain of 780 accessions and wish to verify the map using an introgression-line population of 160 progeny lines grown in two different locations. In this project we investigate lipid-metabolism. Among the candidate genes identified in our GWAS, we will focus especially on the identification and verification of lipases. This project is realized in close collaboration with Prof. Dabing Zhang and Dr. Jianxin Shi (Jiao Tong University, Shanghai, China). (Studied by Dr. Isabel Orf)
In tomato lipids and fatty acids serve as precursors for many volatile compounds that contribute to the fruit's taste. This exacerbates the incentive for the characterization of lipid processes, since lipids, despite their relative low abundance in tomato fruits, are crucial for key agronomical traits in this crop. We are using genetically defined populations of various tomato strains, quantify the levels of numerous lipid species in each strain, and finally correlate the genetic with the lipidomic data to identify loci in the tomato genome that contain genes that control lipid metabolism and regulation. (Studied by Anastasiya Kuhalskaya).
One more crop researched in our group is maize. In collaboration with Profs. Weiwei Wen and Yan Jianbing (Huazhong Agricultural University, Wuhan, China), we are mapping lipids and secondary metabolites in kernels, in order to address the staggering natural variation in this staple crop and to identify genetic determinants for the production of oil and other important nutrients.
