Isotope labeling as a method to annotate and quantify metabolites and proteins

The annotation of metabolites in untargeted metabolomic analysis can be extremely tedious and often produces ambiguous, or even no results. As we and others have shown previously isotope-labelling approaches in combination with complex database searches allow not only for the bona fide distinction of biological from contaminating background peaks, but also increase the reliability of the elemental composition annotation by decreasing the false discovery rate. In order to further reduce ambiguities for the elemental formula annotation, we increased the stringency of our method by including 13C labelling, 15N and 34S metabolically-labelled metabolomes into our analysis. The labelling efficiencies for these isotopes were on average greater than 90%.
Isotope-labelled compounds have, except for their molecular mass, identical physico-chemical properties leading to almost identical chromatographic behaviour. Due to the mass difference of the monoistopic peaks of the different isotopically labelled co-eluting compounds, the absolute number of the labelled elements of a detected molecule can be directly deduced. This principle, and its use for the annotation of elemental compositions, is illustrated in the figure below for a semi-polar compound extracted from Arabidopsis leaf samples.

As shown above, in all four extracts a major peak with a retention time of 4.17 ± 0.02 min can be observed. Zooming into the mass spectra of these peaks shows that they are reproducibly shifted according to the number of carbon (+12 = C12), nitrogen (+1 = N1), or sulphur (+6 =S3) atoms, providing major information for the elemental formula annotation. The missing elements, namely the number of hydrogen, phosphate, and/or oxygen can now, after having fixed the number of the labelled elements, easily be deduced from the accurate 12C monoisotopic mass of the measured compound by using a de-novo elemental formula calculation.
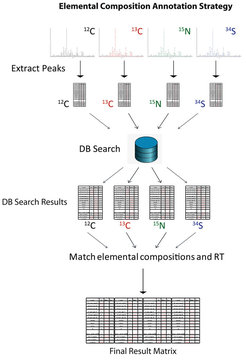
In a next step we develop an automatic, database-dependent strategy for the annotation of elemental compositions of all the peaks in the recorded chromatograms. The strategy of this approach is based on the independent extraction and alignment of all the peaks from the four different isotope-labelled samples (12C (unlabelled), 13C, 15N, and 34S), providing four independent data matrices containing the masses, retention times, and intensity values of each measured peak. In a subsequent step, the masses from the aligned matrices (each isotope labelled sample is used separately) were used to perform independent database searches against four databases (12C (unlabelled), 13C, 15N, and 34S).
Each of these four databases contains the exact masses of each compound calculated from the accurate mass of either the mass of the monoisotopic elemental mass (for the unlabelled samples), while the compound masses within the other three databases (isotope labelled databases) are calculated by using the masses of the stable isotope used for the labelling experiment (13C, 15N, and 34S). The result of these four independent database searches leads again to four matrices containing the measured accurate mass and retention time of each measured peak connected to one or several matching elemental compositions. These four matrices can now, in a final step, be merged by matching the identical elemental compositions between the different isotope-labelled samples and their corresponding retention times.
Depending on the size and the biological relevance of the databases used for these searches (we used KEGG and KNApSAcK for the polar fraction and Target Lipids for the organic fraction), we were able to match 4,908 polar and semi-polar elemental compositions, while 2,392 lipophilic elemental compositions were assigned.


