Subcellular distributions of metabolites and proteins within the cell: Non-aqueous fractionation (NAF)
While major improvements have been made in developing methods to analyze both primary and secondary metabolites, there are still major gaps in our understanding of the plant metabolome as a total. This is especially true with regards to the subcellular localization of metabolites and their exchange between subcellular compartments. In order to address this question, we have used an optimized non-aqueous fractionation protocol on Arabidopsis leaves in conjunction with state-of-the-art mass-spectrometry-based metabolite profiling to perform both targeted and untargeted metabolome analyses.

By using a strict statistical validation, the obtained metabolite data were assembled into distinct and robust subcellular distributions anchored to three main cellular compartments: the cytosol, vacuole, and plastid. The compartmental distributions were validated by previously described subcellular localizations of annotated (known) metabolites.
In a further step we made use of the metabolic distribution within the cell and assembled of a topological map of the compartmentalized Arabidopsis leaf cell metabolome, which should allow facilitating hypothesis-generation based on unexpected subcellular distributions of annotated as well as structurally unknown metabolites. While this map only reflects a physiological snapshot of the underlying dynamic metabolic and biochemical flexibility of the plant leaf, it indicated that numerous metabolites could not be explained by the three-compartment model, either due to transport processes or not resolved compartments. Nonetheless, this study provides a broad survey of the fractionated leaf metabolome, establishing a basis for monitoring metabolite localization and transport processes in the plant cell.
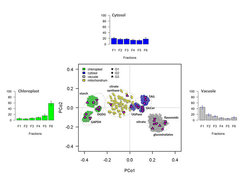
Non-aqueous fractionation of Arabidopsis leaf material facilitated a clear separation of three cellular compartments, which enabled us to reconstruct subcellular metabolite distributions using a marker-based approach. Even though subcellular distributions were precisely estimated using least square approaches, some analytes showed fractions abundances which are not encompassed by the selected markers and thus, their subcellular distributions are only insufficiently explained. While marker-“free” reconstructions showed the existence of unconsidered compartments or intermediate clusters, neither a biological relevance could be attached, as they are mainly comprised of structurally unknown analytes, nor could an unambiguous designation for these clusters be determined. Currently, this represents one of the main limitations in NAF studies, as even the subcellular localization of structurally identified (known) metabolites are often not described in literature and even then, their localization might still be variable. Therefore it is clear that a comprehensive framework of marker distribution to align and assemble metabolites needs to be established based on the measurement of known unambiguously localizable compounds. This criterion seems to be so far only covered by proteomics and thus, a proteomics approach on the individual NAF fractions would be probably the best way to approach this. With proteomic data, especially membrane localized proteins, in combination with the molecular and metabolite markers, a more holistic and precise overview of the dynamics of the metabolome can be determined as the location of many unconsidered compartments can be possibly delineated.

