UPLC-based lipid profiling
The identification and annotation of the lipids is obtained by their specific fragmentation behavior, which is usually measured in highly sensitive triple Quadrupole (QqQ) mass spectrometers. The main disadvantage of this approach lies in the fact that only lipids with an annotated fragmentation behavior (targeted analysis) can be measured, while many unknown compounds (untargeted analysis) are ignored in these studies. To partially overcome this problem, we established an UPLC-FT MS platform that allows to not only extract expected or known lipids, but also to compare and relatively quantify eluting compounds of as yet unknown identity.
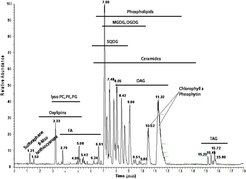
On the contrary to the shotgun lipidomic approach, the UPLC-MS-based strategy does not necessarily rely on the MS/MS data, even though it can be used to confirm the identification of a measured lipid. The main characteristics of the annotated lipids required for the proper identification are their mass (m/z) and their retention time. Confidence in the annotation of a measured compound can be increased with the number of parameters that this compound shares with related compounds. Since lipids are constructed as modular molecules and usually vary only slightly between different species within a lipid class (extension of the fatty acid chain length or the degree of saturation), they have a very systematic mass and retention time behavior. Therefore, both these parameters allows the validation of lipids within a specific class by simply plotting the m/z and RT values of the measured species of the most abundant adduct in a scatter plot.
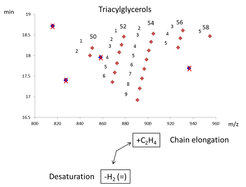
Accordingly, the lipids with longer fatty acid chains lead to a higher mass and increased retention time, while fatty acids with higher degrees of un-saturation result in lipids with lower masses and decreased retention times. As a consequence, a diagonal series appears within the plots. These contain lipid species with the same number of carbons atoms in the fatty acid chains but show decreasing number of double bonds from left to right. Wrongly annotated or unusually distributed lipids can be easily detected within these patterns since they appear as dots outside the systematic scatter pattern.

